Hybridization between two sympatric ranid frog species in the northern Sierra Nevada
Ryan A. Peek, Mallory Bedwell, Sean O’Rourke, Greta M. Wengert,
Caren Goldberg, Michael R. Miller
 ]
]
As freshwater systems and biota are rapidly declining globally, conservation efforts will require assessment of the adaptive capacity of populations to rapid environmental change.
Small populations with limited genetic diversity may have reduced adaptive potential and difficulty responding to future environmental change.
Acknowledgements
Center for Watershed Sciences & colleagues
Corey Luna, Rick Wachs, and Sarah Mussulman
Thanks to Jamie Bettaso, Mourad Gabriel, Cathy Brown, Colin Dillingham, Tina Hopkins, Neil Keung, and WSU and USFS field crew members

Hybridization
Hybridization
Hybridization may be an important evolutionary mechanism (homogenization, speciation, adaptation)
Hybridization
Hybridization may be an important evolutionary mechanism (homogenization, speciation, adaptation)
Hybridization between closely related taxa can promote gene flow (introgression), or combine parental genotypes in ways that yield reproductively sterile or isolated lineages
Hybridization
Hybridization may be an important evolutionary mechanism (homogenization, speciation, adaptation)
Hybridization between closely related taxa can promote gene flow (introgression), or combine parental genotypes in ways that yield reproductively sterile or isolated lineages
Hybridization events may be short-lived and difficult to detect, and are generally considered rare in vertebrates
- Cross-breeding between species, or hybridization, in vertebrates has generally been considered a rare occurrence.
- Hybridization may be an important evolutionary mechanism for either homogenization (reversing initial divergence between species), speciation (from reproductive isolation of hybrid populations), or adaptation (transfer of adaptive alleles) (Abbott et al., 2013; Barrera-Guzmán, Aleixo, Shawkey, & Weir, 2018; Mallet, 2007).
Can Rana sierrae (RASI) and
R. boylii (RABO) hybridize?
 Sierra yellow-legged frogs (RASI)
Sierra yellow-legged frogs (RASI)
 Foothill yellow-legged frogs (RABO)
Foothill yellow-legged frogs (RABO)
Can Rana sierrae (RASI) and
R. boylii (RABO) hybridize?
 Sierra yellow-legged frogs (RASI)
Sierra yellow-legged frogs (RASI)
 Foothill yellow-legged frogs (RABO)
Foothill yellow-legged frogs (RABO)
Probably not based on Zweifel's work (1955)
- Unlike other ranid frog species with broad areas of potential intergradation (Shaffer et al 2004) RASI/RABO rarely occur sympatrically
- breeding experiments by Zweifel (1955) found high incidences of embryological abnormalities—indicating a post-zygotic barrier between the species (low viability in fertilization).
- However, these experiments only crossed female R. sierrae with male R. boylii from very different sites (Butte and Nevada County vs. Contra Costa County).
Management Challenges:
Species have very similar morphology & habitat preferences Zweifel (1955)
Management Challenges:
Species have very similar morphology & habitat preferences Zweifel (1955)
Detecting hybrids & assigning individuals to species in overlapping zone can be difficult, and imprecise using field identification methods
Objectives:
Objectives:
Use RADSeq/Rapture to look for hybridization and identify species
Estimate migration rates between species across 3 watersheds
Revise "map" to help USFS with management/conservation objectives
- Hybridization can promote gene flow (introgression) between species
- Important evolutionary mechanism for:
- homogenization, adaptation, or speciation
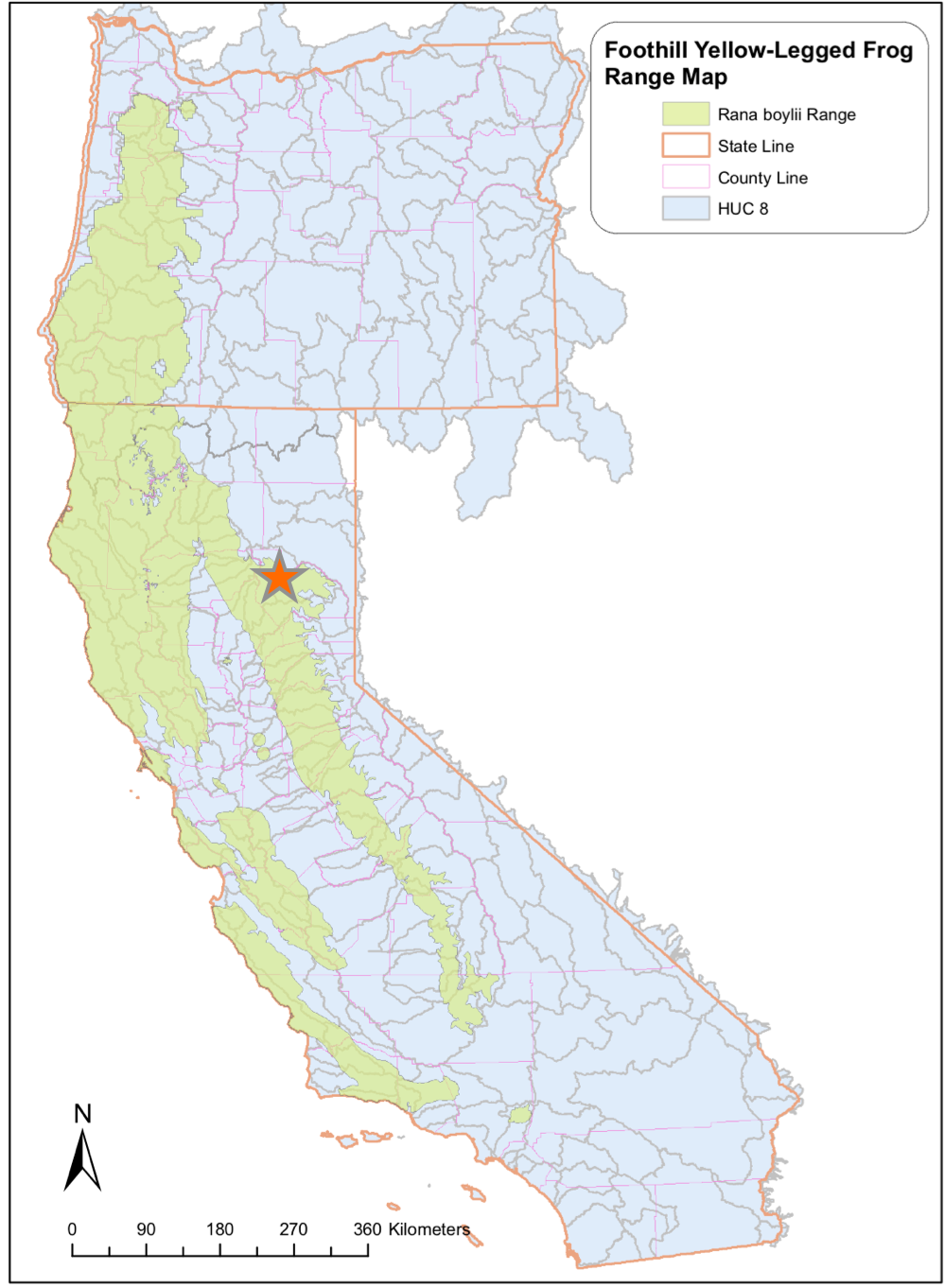
Foothill yellow-legged frogs (Rana boylii)
Foothill yellow-legged frogs (Rana boylii)
Obligate river breeding frog, uses wide range of habitat (<1500m), but has disappeared from over 50% of historical range
Being evaluated as candidate for state and federal listing under ESA
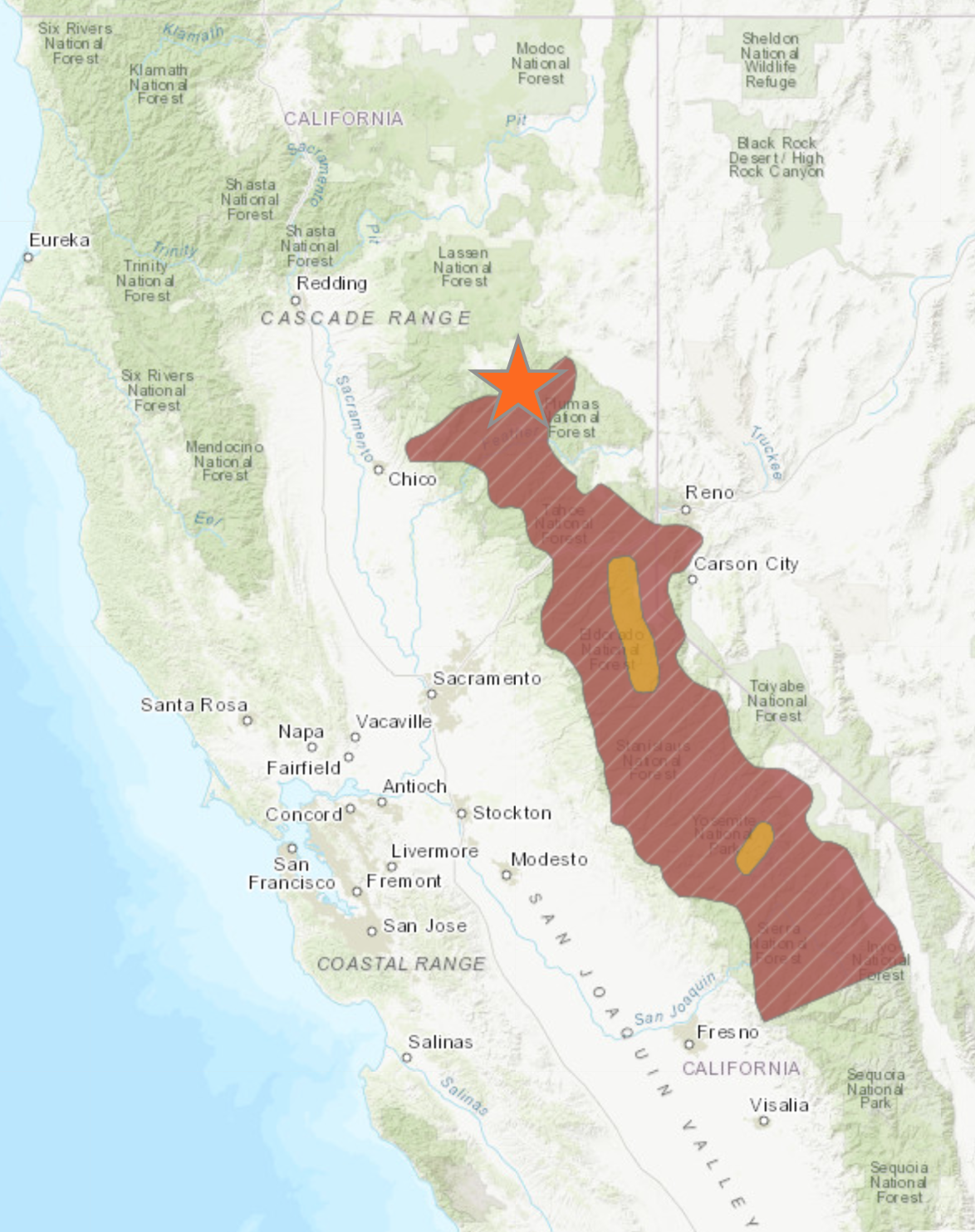
Sierra yellow-legged frogs (Rana sierrae)
Sierra yellow-legged frogs (Rana sierrae)
Federally listed species, can use wide range of lotic/lentic habitat (~1,500 - 3,660m)
Northern boundary of the species range is largely in the Feather basin
Methods: RADSeq & RAD-Capture (RAPTURE)
genome-wide, targeted genotyping approach (Ali et al. 2016)

Flexible and cost efficient, thousands of samples multiplexed at once (high throughput).
- Many amphibian species cryptic/lack data, Large genome size, assessment and monitoring hard to do
- restriction site-associated DNA sequencing, highly efficient genotyping across thousands of individual samples for targeted loci
- For frogs, can sample with non-invasive methods using mouth swabs, or tadpole tail clips
- (i.e., small pops. with limited connectivity, low diversity)
- RAPTURE/RADSeq is a powerful and effective method
RAD Tags = sequences downstream of restriction sites. In order for samples to be sequenced on an Illumina machine, after cutting fragments must be ligated to adapters that will bind to an Illumina flow cell (RADSeq uses modified Illumina adapters that enable the binding and amplification of restriction site fragments only)
- Digest with Sbf1
- Add RAD adapters
- Sonicate/shear
- purify
- specify Capture baits/loci of interest (better/more sequencing at locations you want to look at)
- Different cutters at different frequencies in the genome, this is rougly 300 cut sites per 100,000 bp
PCR duplicates can be identified as fragments that are identical in insert length and sequence composition, because random shearing ensures that fragments at a given locus are unlikely to be of equal length unless they are duplicates (Fig. 1; Davey et al. 2011; Hohenlohe et al. 2013). In contrast, for methods without a random-shearing step, all fragments at a given locus are expected to be of equal length whether or not they are PCR duplicates. Pooled DNA was sheared in a Bioruptor NGS sonicator.
Frog Samples & Sequencing
- 1,443 individual Rana frogs from 17 species
Frog Samples & Sequencing
- 1,443 individual Rana frogs from 17 species
- Designed ~8,500 Rapture baits (“tags”)
Frog Samples & Sequencing
- 1,443 individual Rana frogs from 17 species
- Designed ~8,500 Rapture baits (“tags”)
- Paired end sequence data from Illumina HiSeq
Frog Samples & Sequencing
- 1,443 individual Rana frogs from 17 species
- Designed ~8,500 Rapture baits (“tags”)
- Paired end sequence data from Illumina HiSeq
- de novo assembly via Novoalign and PRICE assembler, ANGSD for analysis
Sequencing Results
- Samples collected across 56 different localities in 3 watersheds
Sequencing Results
- Samples collected across 56 different localities in 3 watersheds
- Filtered & retained 311 samples that contained >25,000 alignments, with mean alignments per sample = 229,485
Sequencing Results
- Samples collected across 56 different localities in 3 watersheds
- Filtered & retained 311 samples that contained >25,000 alignments, with mean alignments per sample = 229,485
- Mean number of samples per site = 8
Sequencing Results
- Samples collected across 56 different localities in 3 watersheds
- Filtered & retained 311 samples that contained >25,000 alignments, with mean alignments per sample = 229,485
- Mean number of samples per site = 8
- Kept all sites regardless of samples/site
Hybridization Sampling Sites
Bean Creek/Spanish Creek largely the only locations where both species have been documented together, however, additional samples from Slate Creek and a few other small tributaries may show both spp (Goldberg/Mallory's MS)
All but 2 individuals were assigned to species using PCA
All but 2 individuals were assigned to species using PCA
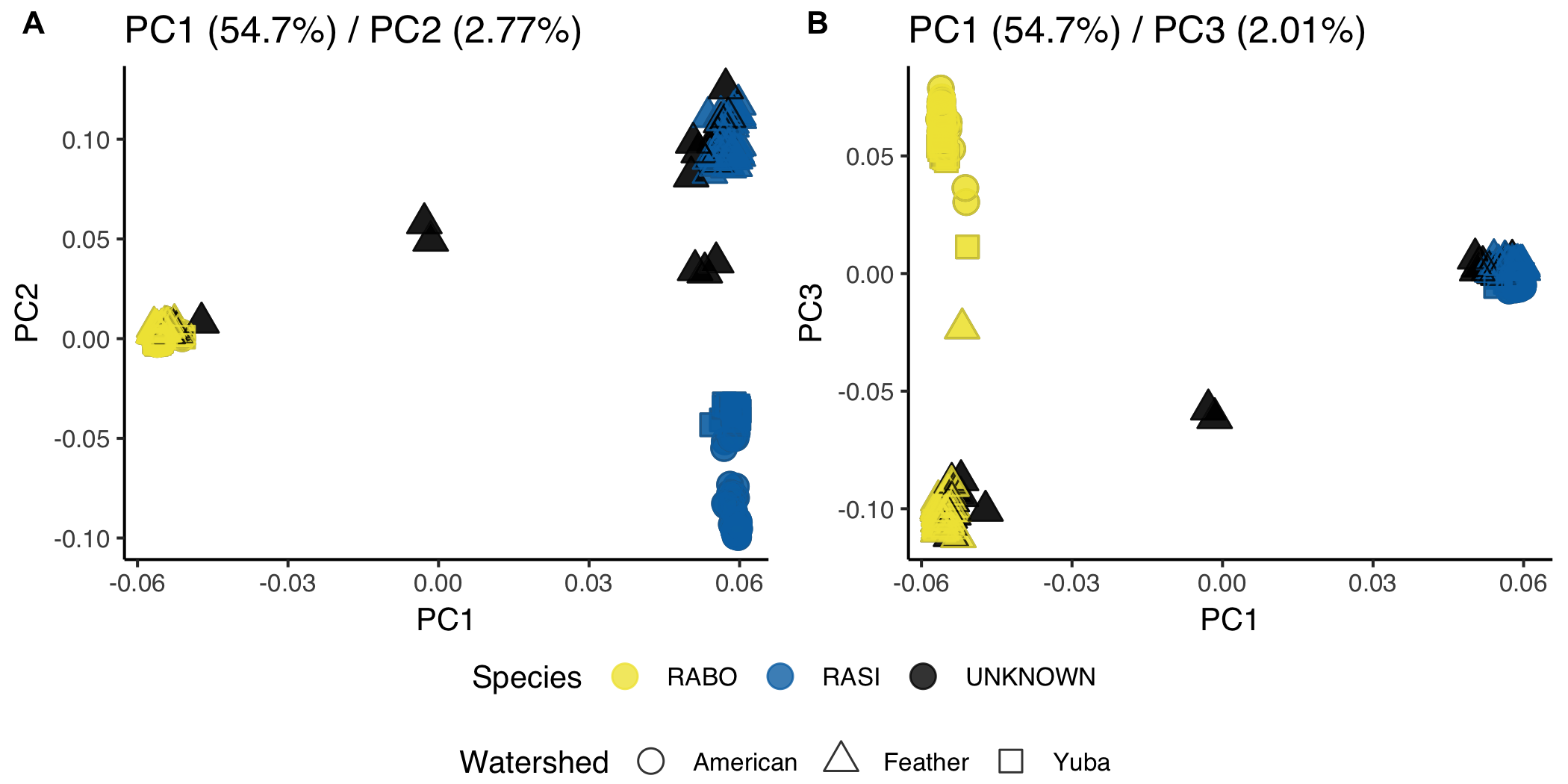
Strong pattern of divergence between the species
Admixture
Admixture

- Used NGSAdmix to assess population structure and individual ancestry from genome-wide SNPs
- Admixture showed the same two unknown samples from the Feather basin were had approximately 50% ancestry from each species.
- ancestry in the individuals designated as “unknown” in the field also showed very low levels of mixed ancestry between the species
- very low or nearly non-existent levels of mixed-ancestry in the American and Yuba basins as compared to the Feather. However, introgression between spp appears asymmetric, with a greater proportion of ancestry from R. boylii occurring in the R. sierrae samples
F1 vs. F2 Test on hybrids
Tested whether the hybrids were F1 (first-generation) or F2 (progeny of two F1’s)
F1 vs. F2 Test on hybrids
Tested whether the hybrids were F1 (first-generation) or F2 (progeny of two F1’s)
Identified putative species diagnostic SNPs (n=3,062)
F1 vs. F2 Test on hybrids
Tested whether the hybrids were F1 (first-generation) or F2 (progeny of two F1’s)
Identified putative species diagnostic SNPs (n=3,062)
Expect F1 hybrids to be heterozygous at all SNPs,
F1 vs. F2 Test on hybrids
Tested whether the hybrids were F1 (first-generation) or F2 (progeny of two F1’s)
Identified putative species diagnostic SNPs (n=3,062)
Expect F1 hybrids to be heterozygous at all SNPs,
Expect F2 hybrids to be heterozygous for at 50% of SNPs and homozygous at the remaining 50% with 25% assigned to each species
F1 vs. F2 Test
Our filtering process (see Methods) yielded 3,062 putative diagnostic SNPs that were homozygous for different alleles in R. sierrae or R. boylii samples and also had successfully called genotypes in the two hybrid individuals
- We observed extremely high heterozygosity and very low homozygosity (6% genotyped as R. boylii, 4% R. sierrae, and 89% were heterozygous) (Figure 4). This level of heterozygosity is far greater than expected for F2 individuals
In addition, both hybrid individuals had RASI mitochondrial DNA (Bedwell 2018), indicating the maternal ancestor was from a RASI individual and the paternal ancestor was RABO
- non Hz SNPs may be expected (at low levels), due to low coverage sequencing data; genotyping from low coverage sequencing will cause a low frequency of erroneous homozygous calls, because only one of the two alleles is sampled, causing heterozygotes to be called as homozygotes
Hybrid Locations
Demographic Modeling
Demographic Modeling
Used
fastsimcoal2(Excoffier & Foll, 2011) coalescent simulations to estimate migration rates between species within each watershed
Demographic Modeling
Used
fastsimcoal2(Excoffier & Foll, 2011) coalescent simulations to estimate migration rates between species within each watershedUsed all individuals except the two hybrids to calculate migration probability (or the per generation likelihood that any gene from one population transfers to another)
Demographic Modeling
Demographic Modeling
Migration rates highest in Feather compared to Yuba and American
Demographic Modeling
Migration rates highest in Feather compared to Yuba and American
Divergence time estimated at 371,000 generations or ~742 kya to 1.1 mya if using generation time of 2-3 years
The best model based on maximum likelihood estimated the time since divergence between R. sierrae and R. boylii was 370,856 (370,041–371,670) generations.
Early Pleistocene began ~1.8 mya ("Great Ice Age").
Summary:
Strong divergence & limited hybridization detected between RASI and RABO, however F1 hybrids were identified in Bean Creek
Summary:
Strong divergence & limited hybridization detected between RASI and RABO, however F1 hybrids were identified in Bean Creek
Demographic modeling suggests migration rates between species are highest in the Feather Basin, and divergence estimates coincide with onset of Pleistocene Ice Age
Summary:
Strong divergence & limited hybridization detected between RASI and RABO, however F1 hybrids were identified in Bean Creek
Demographic modeling suggests migration rates between species are highest in the Feather Basin, and divergence estimates coincide with onset of Pleistocene Ice Age
Hybridization outcomes that fail to produce successful offspring (sterile F1 hybrids) may have a greater cost on the species with low numbers of effective breeders (i.e., RASI), something to consider under future scenarios (climate change, population crash, etc)
unlikely that there is currently the potential for major introgression between R. sierrae and R. boylii, particularly as hybridization initially may not be adaptive and is often selected against (Abbott et al., 2013; Streicher et al., 2014).
hybridization outcomes that fail to produce successful offspring (sterile F1 hybrids) may have a greater cost on the species with low numbers of effective breeders, affecting both locally adapted populations and negatively impacting the probability of population persistence in a given region
under future scenarios (e.g., warming climate, range contraction, population crashes) the loss of even several breeding individuals (via reproduction with R. boylii) may have a significant impact in declining populations
"The good life of any river may depend on the perception of its music; and the preservation of some music to perceive. Aldo Leopold"